Here I discuss my attempt at applying Computer vision & Deep learning techniques to detect endocrine & other colon related cancer: BE,suspicious, HGD, Cancer, polyp.
Endoscopy is a widely used clinical procedure for the early detection of cancers in hollow-organs such as oesophagus, stomach, colon and bladder. Computer-assisted methods for accurate identification of tissues that are malignant, or potentially malignant has been at forefront of healthcare research to improve clinical diagnosis. Many methods to detect diseased regions in endoscopy have been proposed, however, these have primarily focussed on the task of polyp –deleterious intestinal protrusion in the gastrointestinal tract– detection on datasets that lack richness.
This was a challenge organised by the University of Oxford, the organisers consolidated data sets from various leading research institues on cancer to enrich data variety inorder to minimise artefact errors– errors inevetibly occuring during scientific experiments– , and made it public to encourage AI enthusiasts to implement solutions to push the frontiers of sophisticated cancer diagnosis on the enriched data set.
The problem was a segmentation challenge where one is expected to delineate regions of interest in images, in our scenario disease infected regions, which belonged to either of 5 classes.
The data set, although was diverse in aspects of variety, the data set quantity was scanty(about 386 images), which is way too low to apply deep learning techniques.
This required implemeting data augmentation techniques to increase the size of data, along with standardising the data set like resizing unequal shapes, removing blurs etc..
For this challenge I implemented ResU-Net, a combination of the popular U-Net & Residual net architechtures from the family of Convolutional Neural Networks(CNN), & scored a decent score; also I tried the more advanced version of ResU-Net, the ResU-Net++, which did not improve the score much.
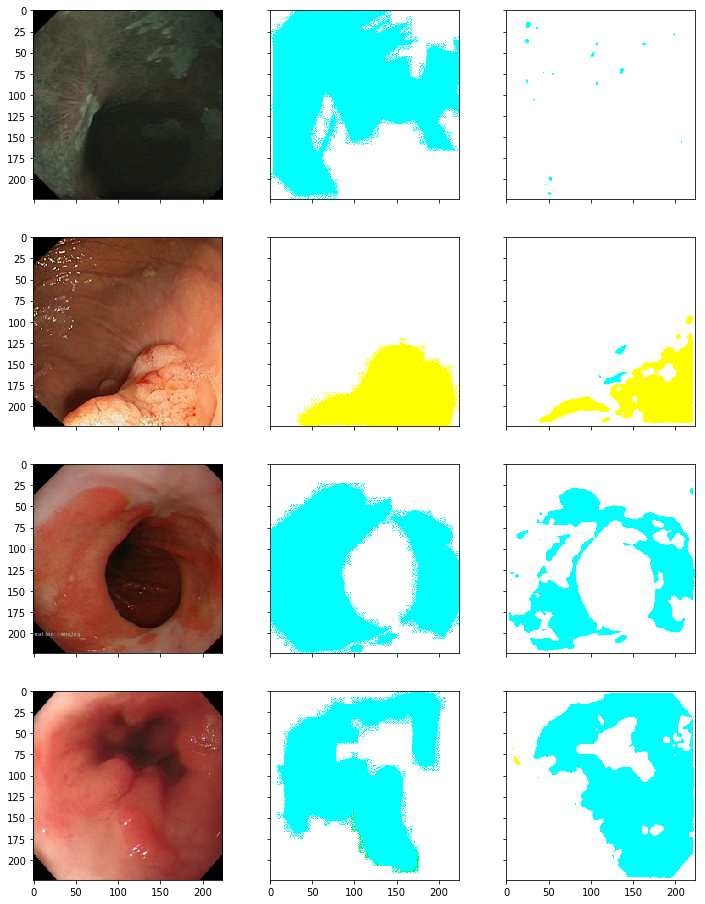
However, trying training hacks to improve training like training the images in two phases, each with different size of image data set certainly improved the score.
From the results, it seems that my model is correctly predicting the disease(color), however it needs to improve in predicting the regions affected(outline); also it was observed that this often happens with diseases other than BE(cyan) & polyp(yellow), perhaps because data regarding other diseases (suspicious,cancer,HGD) is less as compared to BE & polyp.
Please refer to the work over here.